Semantic Data Integration with RDF
Автор: Konstantin Pentchev, Vassil Momtchev
Источник: Тhe 7th Framework Programme for Research and Technological Development.
Introduction
This chapter presents an analysis of the different levels of performing data integration. Ziegler and Dittrich [1] define multiple integration levels depending on its specificity. They start from
1) Manual integration – no real integration is done since the interpretation is performed by the end-user;
2) Common user interface – data from relevant sources are displayed in a single view in the application;
3) Integration by applications or middleware –integration is done on the concrete application level where the developers are relieved only from implementing common integration functionality;
4) Uniform Data Access –information integration is realized by virtual data or data abstracted from its physical structure in runtime;
5) Common Data Storage – is the physical reorganisation or replication of the existing data to a new place and possibly new global schema.
It is a general rule that the integration becomes more efficient when it is moved closer to the physical storage. Thus, when we need to operate with very large amounts of information like in the context of the Khresmoi project, our choice for efficient data integration is practically limited to the data consolidation (or warehousing) and federation approaches. In the next paragraphs we summarize the different trades-offs in the two approaches and their impact on the knowledge engine design.
Data warehousing is the process of centralizing the information into a common physical storage model. It requires the reorganization and consolidation of all data into a global schema, and may either fully replace the old databases or replicate the information on a regular basis. Either way, data consolidation requires the design and execution of extract transform and load scripts that need to resolve the structure and semantic heterogeneities between the source and the global schema during data loading.
Data federation utilizes a different approach, using the Uniform Data Access level described in [1], which nowadays is often referred to as data virtualisation. Data virtualisation is a technique for abstracting the information from its physical storage and organisation. This enables the cross-data source mediation between the multiple results and query formats during the execution of every request. Thus, all structure and semantic heterogeneities need to be resolved at runtime, which adds efficiency overheads. Furthermore, the pure federation approach is well known for its inability to efficiently deal with many remote join operations. A remote join is the computational merging of information between two distributed physical and/or logical interfaces.
In the Khresmoi project the integration strategies of data warehousing and federation demonstrate significant trade-offs because of the extreme amounts of semantic enabled data that need to be processed efficiently. Calabria [2] extends the two previously mentioned approaches to four architecture approaches presented in Table 1:
Table 1: Different data integration architecture used by the industry.
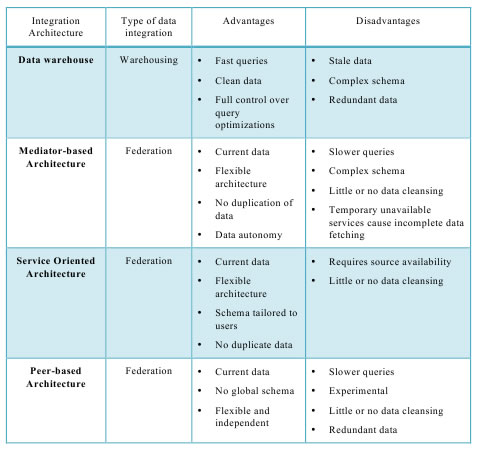
Comparing the different approaches in Table 1, we see that each integration architecture offers significant advantages and disadvantages. It shows that the best solution for processing large-scale information by the KE tends towards warehousing because it provides:
- Forward chaining inference and materialization of trivial implicit statements;
- Even and predictable query performance across all information;
- Unlimited capabilities for data cleaning;
- Integration of consistency checking, which is often critical for the biomedical domain;
The federation approach and the Mediator-based Architecture enable the integration of autonomous data sources that cannot be replicated in a warehouse, because their content is updated too often and/or because of security restrictions. The Peer-based Architecture offers excellent flexibility, but the existing disadvantages, such as the lack of a global schema and the slow querie times.
RDF Warehousing
An RDF warehouse requires the translation of all data sources to RDF triples and loading the statements into different named graphs (contexts). Keeping different dataset in separate named graphs guarantees the minimal provenance information required in order to support incremental information updates.
The Linked Life Data (LLD) service [3] is an example of an RDF warehouse project that demonstrates excellent performance for a wide range of SPARQL queries against billions of RDF statements. The service relies on a highly efficient persistence of RDF, a query optimizer and an integrated forward chaining reasoner that enables the indexed search of implicit statements. Once all the information is consolidated into a single physical structure, resource alignment rules are defined to link related identifiers. Figure 1 depicts six alignments rules, where the dashed lines and the blue text of the captions (used either as part of the URI or literals) designate the criteria for linking the information. Since the specified mapping rules are not universally applicable for arbitrary RDF datasets, they are manually controlled for each specific subset.
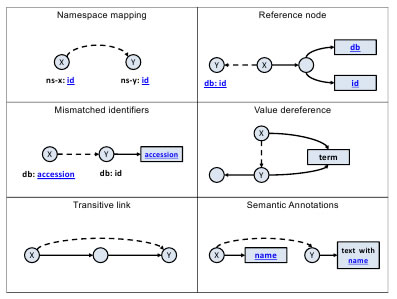
Figure 1 Resource alignment patterns in LLD.
Figure 2 illustrates the semantic aspect of the instance mappings.
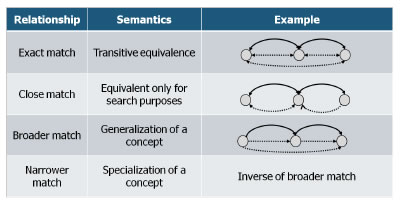
Figure 2: Instance mappings in LLD using the SKOS vocabulary.
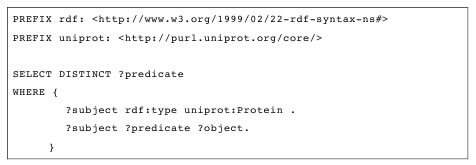
Local RDF storage engines can provide full control over the query optimisations and statistics on each value's associations, allowing the calculation of optimal execution plans for complex information joins. Queries with unbound predicates are especially difficult to optimize. The SPARQL query presented in Table 2 lists all unique predicates for resources of type Protein. The first pattern executed against LLD 0.8 results in 16,505,340 possible bindings. The second pattern to be merged with the first one results in 5,120,886,447 possible bindings. This yields a total of 8.45E+16 tuples if naive optimisation is used. However, the total execution time for the presented query is less than 60 seconds despite its extreme complexity. Similar types of queries are not practical for any sort of pure federated environments because of the previously mentioned remote join limitation.
Table 2: SPARQL query with unbound predicate.
In conclusion, warehouse architectures, like the one implemented by the LLD project, guarantee fast queries because they offer a single data model, storage engine-provided query optimisation strategies and the possibility of data quality control and cleaning. However, the central control over the information comes at a cost. Every warehouse has redundant and stale data, which requires regular updates. In some scenarios, the warehousing approach is not compatible with specific data source licences or imposed security restrictions. The next chapter will investigate how the KE architecture may overcome these limitations.
SPARQL Endpoint Federation
The service-oriented and mediator-based architectures are very similar, with the exception of the API interface that is used to communicate with the federated databases. In the service-oriented architecture the integration middleware will support any type of data objects returned by the service and will automate their mappings to the global model. In the context of RDF, the mediator-based architecture is very close to the SPARQL 1.1 federation working draft [4]. The working draft is produced as a response to the increasing number of public SPARQL endpoints, the need to integrate information distributed across the web and to overcome the licence or security restrictions imposed by specific data sources. The SPARQL language syntax and semantics are extended with the SERVICE and BINDINGS keywords that enable the query rewrite to compose a query that delegates specified triple patterns to a series of services.
The SPARQL federation is a very promising approach to the simple integration of very large non-semantic databases. The R2RML working draft [5] presents a language for expressing customized mappings from relational databases to RDF dataset that could be further exposed as a virtual SPARQL endpoint over the mapped relational data. Still, a major challenge for efficient query execution is the limited interface that does not include the sharing of any statistics to be used in the optimisation. Nonetheless, the access to virtual SPARQL endpoints is a practical way to overcome security and licence limitations.
[1] Ziegler, Patrick & Dittrich, Klaus R.: Three decades of data integration - All problems solved? Kluwer (2004), S. 3-12.
[2] Calabria A., Data Integration for Clinical Genomics, Doctorate Thesis, 2009, available at: http://boa.unimib.it/bitstream/10281/19219/3/Phd_unimib_716358.pdf
[3] Vassil Momtchev, Deyan Peychev, Todor Primov, Georgi Georgiev. Expanding the Pathway and Interaction Knowledge in Linked Life Data. In Proc. of International Semantic Web Challenge, 2009.
[4] Prud'hommeaux E., Seaborne A., SPARQL 1.1 Federation Extensions, W3C Working Draft 1 June 2010
[5] Souripriya Das, Seema Sundara, Richard Cyganiak. R2RML: RDB to RDF Mapping Language W3C Working Draft 24 March 2011