Watershed Segmentation Algorithm for Medical Confocal Image Analyses Towards In Vivo Early Cancer Detection
Man Kin Derek Ho
Department of Biology, Johns Hopkins University
NNIN REU Site: Microelectronics Research Center, The University of Texas at Austin
NNIN REU Principal Investigator: Dr. John X. J. Zhang, Biomedical Engineering, The University of Texas at Austin
NNIN REU Mentor: Karthik Kumar, Electrical Engineering, The University of Texas at Austin
Contact: derekho@jhu.edu, john.zhang@engr.utexas.edu, kkumar@mail.utexas.edu
Abstract
This REU project successfully demonstrated an automated image segmentation technique to overcome artifacts from in vivo images and provide real time, accurate analysis of nuclear size, density, and nuclear-cytoplasmic ratio, critical visual markers of epithelial precancers. The algorithm was first calibrated using optical images of microfluidic droplets, and then applied on confocal images of oral cavity tissues. All images were segmented successfully to provide accurate count (95% with 6.2% standard deviation) of cells or droplets.
Introduction
6.5 million deaths annually. Although widely considered a disease of the developed world, 60% of cancers occur in developing countries, where low per-capita healthcare expenditure, unreliable infrastructure and facilities render advanced cancer screening technologies inaccessible. In lieu of these factors, we have developed a low-cost, handheld, microelectromechanical systems (MEMS)-based in vivo confocal microscope for sub-cellular-resolution imaging of tissue towards early detection of epithelial precancers from which 85% of cancers originate. Currently, endoscopic procedures are performed for biopsy samples and images are manually segmented for initial testing of pre-cancer. However, this results in long turnaround time, high costs, discrepancies among different segmentation methods, and
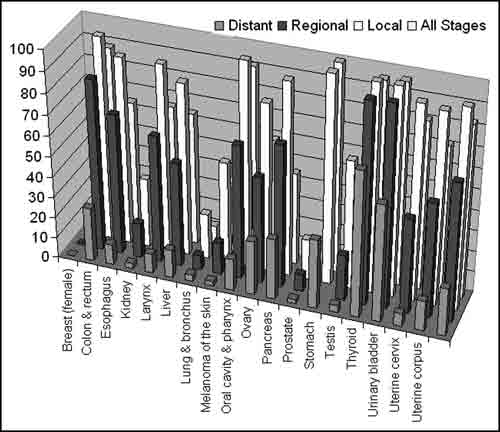
Figure 1: Statistics showing survival rates vs. cancer diagnosed at different stages.
inconvenience for patients. An advanced algorithm is therefore needed to provide fast, low-cost, standardized results which are essential for in vivo pre-cancer detection.
Algorithm Outline
The image was imported into MATLAB® and converted to grayscale for faster processing using inbuilt functions. Imclearborder was used to eliminate incomplete nuclei and only account for complete and visible ones. The gradient magnitudes were calculated and the threshold was increased to outline cell membranes.
The normal watershed transform usually over-segments so we used the more advanced marker-controlled watershed transform. Foreground objects were marked with local maxima to ensure only wanted objects were accounted for. Strel and graythresh were used to mark the background in black against the resized white foreground markers. In order to only count minima at
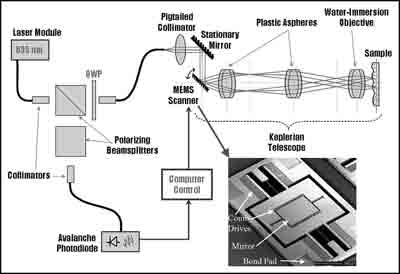
Figure 2: Illustration of our imaging setup with top view of microscanner shown (Inset).
markers and not the entire image, imimposemin was used and the transformation was initialized at this point.
To visualize how the watershed function worked, the image was viewed as a topographical surface. The algorithm started at a minima of each marked object and expanded uni-directionally until it reached an edge of another region. The watershed transform was an accurate way to count and segment marked regions. Bwlabel labels connected components of the image, which allowed numObjects and Regionprops to give numerical statistics about the segmented image. Based on results from previous trials, parameters were set up to remove false counts. All nuclei sizes were put into an array and a mean nuclei size was calculated, together with a histogram displaying the occurrences vs. size.
Algorithm Calibration
The algorithm was first calibrated using optical images of droplets in microfluidic channels. Microfluidic channels were fabricated using polydimethylsiloxane (PDMS) with rapid prototyping. A clean silicon wafer was coated with hexamethyldisilazane (HMDS) to ensure good adhesion between silicon and photoresist. Photoresist SU-8 2250 was applied to produce a master of appropriate thickness and the wafer was spun at 1860 rpm for 30 seconds. Under soft lithography, the wafer was exposed to UV light for 50 seconds and developed for 3 minutes. On developing, features of microfluidic channels are transferred to the resist, and the master fabrication is complete. PDMS was poured onto the wafer after applying Sigma-Cote to reduce adhesion between the PDMS and wafer, cured for 45 minutes at 70°C, and peeled to form channels. The master can be used in this manner to reliably reproduce thousands of channels based on a design.
Water droplets dyed with various colors were injected at different arms of the microfluidic channel. The fusion efficiency was observed when different flow rates were applied and the algorithm was capable to segment and count droplets with 93% accuracy and 8.7% standard deviation. Over-segmentation occurred when many droplets did not fuse together. Based on that, an additional criterion was implemented to minimize inaccurately segmented droplets.
Results and Conclusion
Medical confocal images were used for a more rigorous and robust testing of the algorithm. Images were taken from porcine oral cavities with our confocal microscope. The algorithm provided 95% accuracy and 6.2% standard

Figure 3: Porcine oral tissue. Inset: Epithelium and basal layer.
Figure 4: Segmented image with markers shown using our algorithm.
deviation to hand-segmentation. Although over-segmentation did occur, the mistaken segmented regions were obvious in that they were not nuclei based on size and could therefore be discarded.
The results show that our image segmentation algorithm can analyze images taken in vivo and detect nuclei to a good extent. Our algorithm, together with our hand-held in vivo microscope, can provide real-time, biopsy-free results.
Future Work
Bayesian Classifiers can be implemented to detect specific object shape and size, which allows an observation of different cancer stages and an idea of what types of treatments are needed. A GUI can be written for easier usage and other research groups can benefit from automated image segmentation versus manual counting. Also with each image there are different objects and signals in them, thus performing a Fourier Transform can be useful in image analysis.
Acknowledgements
I would like to thank the NSF, NNIN Research Experience for Undergraduates Program, Dr. Sanjay Banerjee, NNIN Facility, and Zhang Research Lab at UT Austin.
References
[1] K. Kumar, K. Hoshino, H.J. Shin, R. Richards-Kortum and X.J. Zhang, “High-reflectivity Two-Axis Vertical Combdrive Microscanners for Sub-cellular Scale Confocal Imaging Applications”, Proceedings of International Conference on Optical MEMS and their Applications (Optical MEMS ‘06), August 21-24, Montana, USA, 2006.
[2] Brette L. Luck, Kristen D. Carlson, Alan Conrad Bovik, Rebecca R. Richards-Kortum, “An Image Model and Segmentation Algorithm for Reflectance Confocal Images of In Vivo Cervical Tissue,” IEEE Image Proc., V14, #9, pp. 1265-1276, Sep. 2005.