International Journal of Engineering and Advanced Technology (IJEAT) ISSN: 2249 – 8958,
3D Volume Rendering Algorithm
Pravin P Kalyankar, S S Apte
Abstract— This paper presents an analysis of the alg orithms used for generating 3D structures from 2D
Index Terms— Marching Cube, 3D surface reconstructio n, isosurafce, Volume rendering, Medical imaging
I. INTRODUCTION
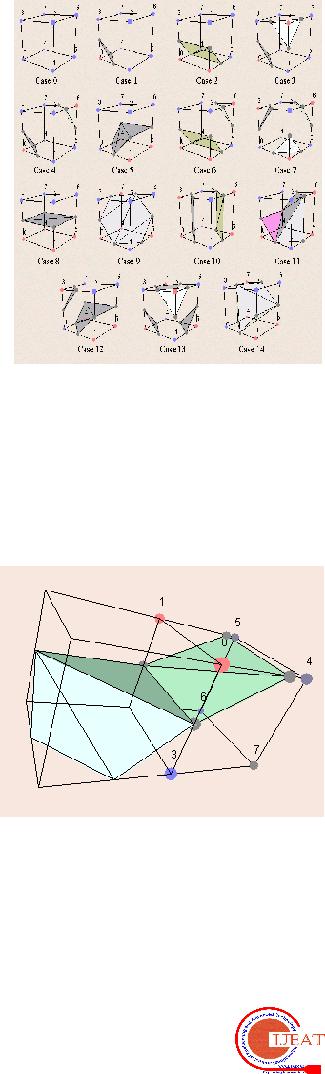
The Marching Cubes (MC) algorithm by Lorensen and Cline [1] is the most popular algorithm for the extraction of isosurface out of volume data. We propose a robust and intelligent triangulation strategy and performing complementary and rotational symmetry operation. This is a significant practical improvement compared to the former scheme of isosurface tilings. The 3D images are developed from 2D CT/MRI scans of a particular object of interest. The core of this algorithm is the creation of 3D CT/MRI rendering software. The software will produce a 3D interpretation of 2D CT/MRI data. Initially, the software stacks the 2D data to form a large cube of data. Statistical classification is used to create and label voxels (3D pixels).
Manuscript received on June, 2013.
Pravin P Kalyankar, Department Of CSE, Dr.BAMU Aurangabad.TPCT COE osmanabad, Indiaity, India.
Dr.S S Apte, WIT Solapur, Department of CSE, Solapur University/ India.
Finally, the Marching Cubes algorithm is used to extract and create polygon surfaces from the volume. Some limited user interaction is permitted such as rotating, scaling and moving the volume.
II.
In order for any 3D surface reconstruction algorithm to properly work, it is important to address the special considerations required of the set of slices for the algorithm to work. Of the different concerns that are normally under consideration for
III. RENDERING 3D SURFACES
Currently, there are two general models used for rendering 3D images. The first is known as Volume Rendering and the second is known as the Marching Cubes Algorithm. Both use voxels (3D square pixels) to determine the 3D area to be constructed. The Marching Cubes (MC) algorithm by Lorensen and Cline is most popular algorithm for extraction of isosurface out of volume data. Our algorithm is adaptive to the small the changes of data or the small changes of the threshold, and obtains more reasonable result of triangulation of isosurface than those produced by standard MC algorithm. Marching Cubes Algorithm is considered a thresholding technique because it uses only the pixel information at the eight corners of the voxel.
IV. MARCHING CUBES ALGORITHM
The current standard algorithm for 3D surface construction is the Marching Cubes algorithm, methods of 3D surface construction. The algorithm implements threshold rendering in order to generate “triangle models of constant d ensity surfaces from 3D medical data” [4]. In summary m arching cubes creates a surface from a three dimensional set of data as follows:
1.Read four slices into memory.
268